Frequently Asked Questions#
Find answers to key questions regarding model usage, functionalities, and troubleshooting to ensure optimal and efficient results.
Note
If you find that you still have unanswered questions, please reach out to us at nci-cmm@mail.nih.gov !
General questions#
Installation issues
If you encounter problems with installing empanada-napari, try restarting napari or installing the plugin using pip. If you still face difficulties, consider creating a new virtual environment to avoid dependency conflicts
I got an “openMP/OMP related error” while working in empanada-napari.
If you get “openMP/OMP related error”, try following these steps:
Try uninstalling pyqt using conda:
conda uninstall -y pyqt
Install and upgrade napari using pip:
pip install napari --upgrade
Install pyqt using pip:
pip install pyqt5
Open napari and try running the empanada-napari modules again.
Note
If this does not resolve the issue, contact us with the error log at nci-cmm@mail.nih.gov
What are the hardware requirements to use empanada-napari?
Empanada-napari can run inference, finetuning, and training modules on GPU and CPU. Though running these processes strictly on CPU will require more time.
Other hardware requirements include:
Operating System: Mac, Linux, or Windows
Python Version: Supported versions are Python 3.9 and below. Later versions (i.e., Python 3.10) are not supported.
GPU Support: Having a GPU installed on your system will significantly increase model throughput, although CPU optimized versions of all models are shipped with the plugin. The plugin relies on pytorch for running models, and GPU drivers must be correctly installed for GPU usage.
Memory: Ensure sufficient memory to handle the processing requirements of deep learning-based image segmentation tasks. 32G should be sufficient.
Storage: Adequate storage space to store datasets, models, and any intermediate results generated during inference or training. 256G should be sufficient.
Why are my denoised images giving me worse results?
MitoNet was trained on images from CEM1.5M that were denoised with histogram equalization, while techniques like noise2void use a distinct denoising method. This variance in denoising techniques causes a significant shift in the characteristics of the data, leading to subpar results when applying MitoNet to denoised images.
Proofreading questions#
Can I undo a proofreading function (i.e., merge, split, morph labels)?
Unfortunately, empanada-napari does not have an undo button. When proofreading or editing segmentations, it is recommended to first duplicate the labels layer in napari. This ensures that you have a reference point and can easily revert to the original if needed.
Tip
You can change the name of the labels layer by double clicking the layer and entering in a new name. This can help you keep track of the changes being made at that time and reduce the need to start over if you accidentally delete a label.
It is also recommended to periodically export segmentations during the proofreading process. This practice minimizes the risk of data loss in case napari crashes unexpectedly, allowing you to resume from the last saved point.
I want to add a new object instance in the labels layer, how do I know what label ID to assign?
There are a few ways to determine available label IDs within the labels layer:
Use the Count Labels module to get the list of already assigned label IDs. Then change the label value in the napari layer controls (see below) and paint in the new instance.
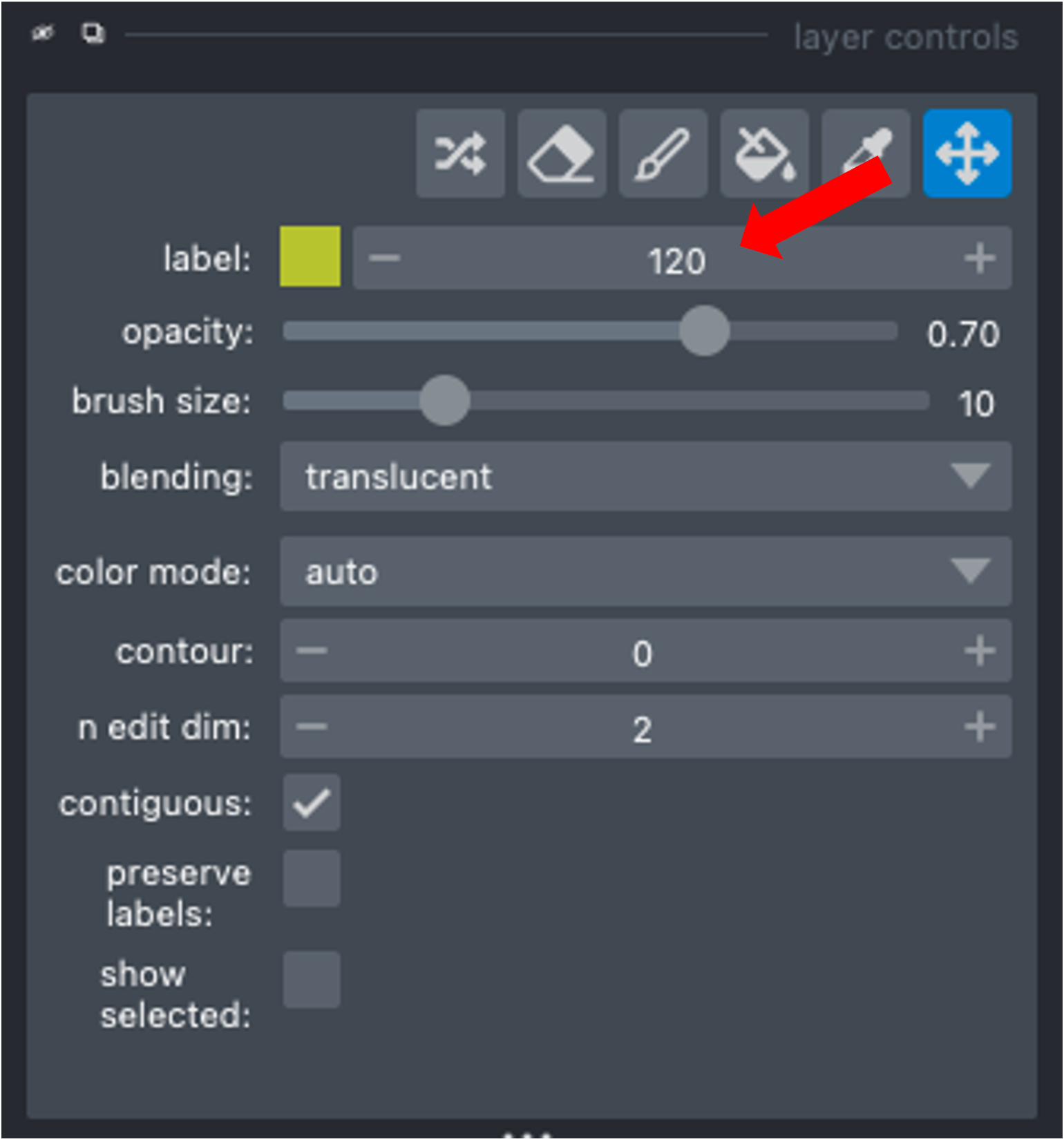
Use the Find next available label module to adjust the napari painter to either append the current list of label IDs (e.g., you have deleted a previous label ID and can now add a new instance with the available label ID) or to add max_label ID + 1.
Training and finetuning questions#
What is the correct file structure needed to finetune and train a model?

There can be multiple name_of_2D_image_or_3D_volume subdirectories. Each must have a subdirectory called images and another called masks. Corresponding image and mask .tiff files must have identical names but reside in the appropriate folder.
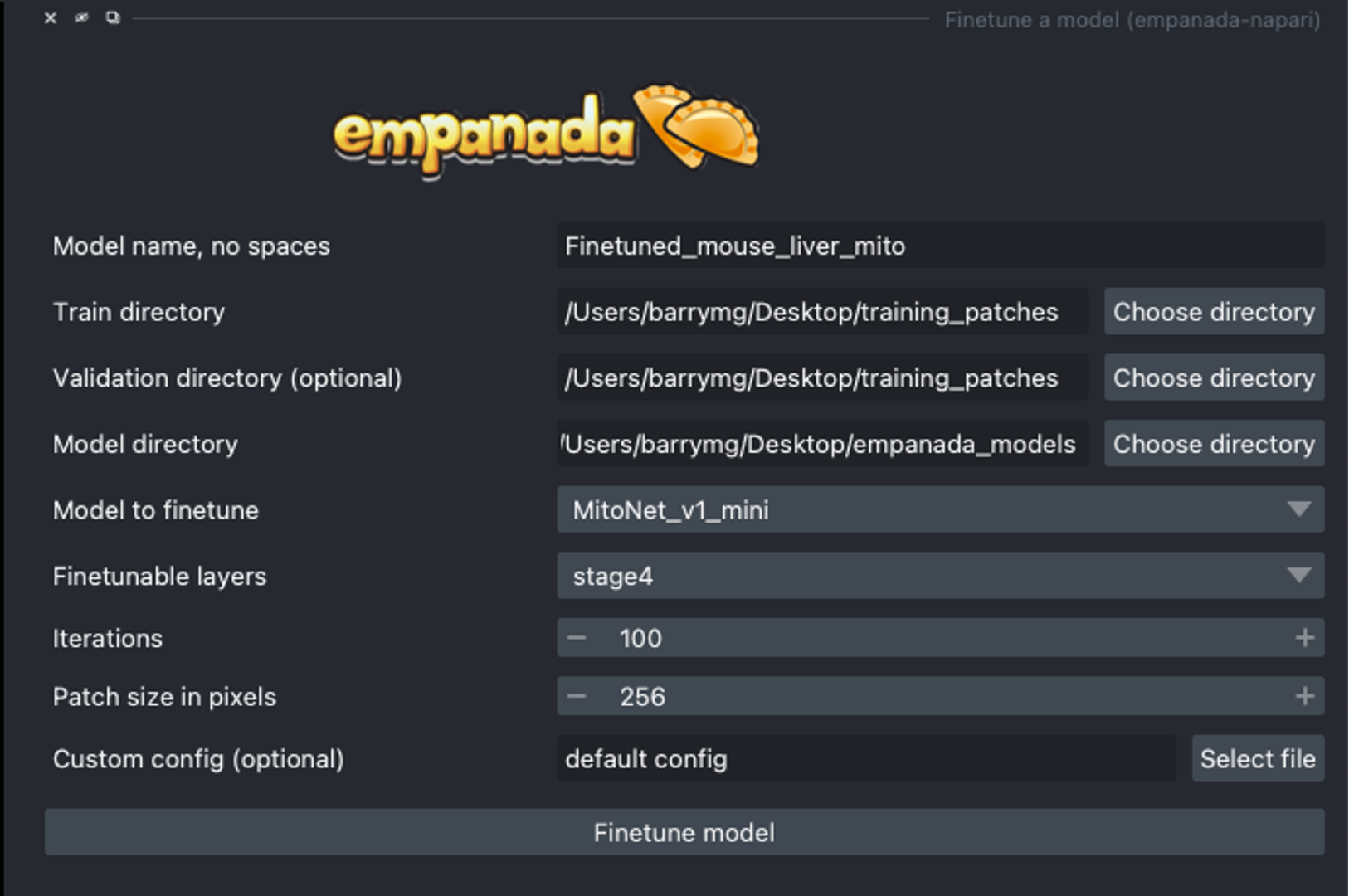
When finetuning a model, what finetunable layers should I select?
When selecting which layers to finetune or unfreeze, consider the following options:
None : Select this option if the model did fairly well during the initial inference on your data.
Stages 1 - 4 : Select between these options depending on how well the model did on the initial inference.
All : Select this option if the model did not perform well on your data. This option will take more time but could offer better results depending on your specific task.
Tip
Experiment with different combinations of frozen and unfrozen layers to observe how it affects model performance. This iterative process can help determine the optimal configuration for your specific task. See Finetuning and Training best practices for more information.
When training a new model, how do I determine the number of iterations?
When training a new model, determining the number of iterations involves a process of testing and optimization. Here are the steps to help you determine the appropriate number of iterations:
Start with a baseline:
A good starting point is around 100 training iterations.
Increasing the number of iterations gradually can help improve the model’s performance.
Optimization:
For a general model like MitoNet, training for more than 500 iterations is usually not necessary unless dealing with a large number of annotated images.
It is recommended to avoid training for more than 10,000 iterations to prevent overfitting.
Finetuning:
Finetuning a model may require different iteration numbers based on the dataset and specific requirements.
If your current dataset is more similar to that of the training dataset, few iterations are needed.
Model Evaluation:
After training, run inference to evaluate the model’s performance to determine if further iterations are needed.
If you find that further finetuning is required, try selecting patches with the Pick finetuning/training patches module of the area the model is struggling with.
Note
Determining the number of iterations can vary depending on the complexity of the training data and the segmentation task. While the above steps can work as as a great starting point, it is recommended to try different configurations to determine the “sweet spot” for your model.
How do I remove a model?
Warning
It is not recommended to delete any other files from this folder besides any user created finetuned model!
To remove an unwanted model that you have either finetuned or trained within empanada-napari:
Go to your files and search for the .empanada folder.
In the config folder you will then find the user created finetuned models.
Select the user created model you wish to delete and delete.
You will need to relaunch napari and empanada-napari to see the changes.
Can I use MitoNet outside of the empanada-napari plugin?
MitoNet is intended for use exclusively within the napari environment through the empanada-napari plugin. This design choice maximizes the model’s effectiveness and ensures that users can take full advantage of napari’s capabilities for their segmentation tasks as well as the proofreading tools developed in the empanada-napari plugin.